Determination of antibody epitope / paratope is essential when establishing the effect of an antibody. Today, most methods for epitope mapping are time consuming and very costly.
Here we are proud to share a high-throughput method offering fast data analysis and is designed for automatization.
We utilized the antibody E10B10 and the antigen murine CD163 for this method development, as they have been proven to bind consistently.
Cluster of Differentiation 163 (CD163), is a surface exposed scavenger receptor on macrophages binding the hemoglobin-haptoglobin complex. It is part of the innate immune response, where it binds both gram-positive and gram-negative bacteria. CD163 is linked to a number of diseases. [1]
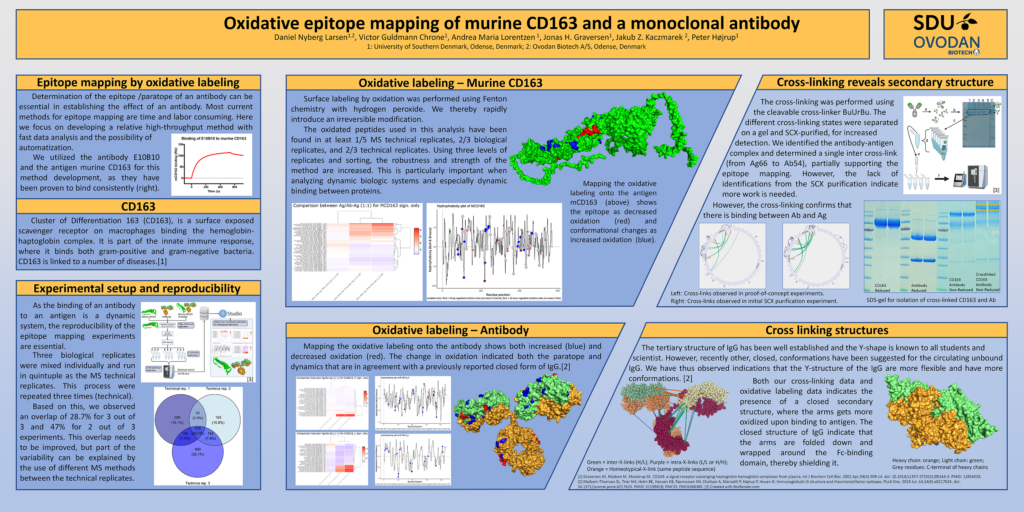
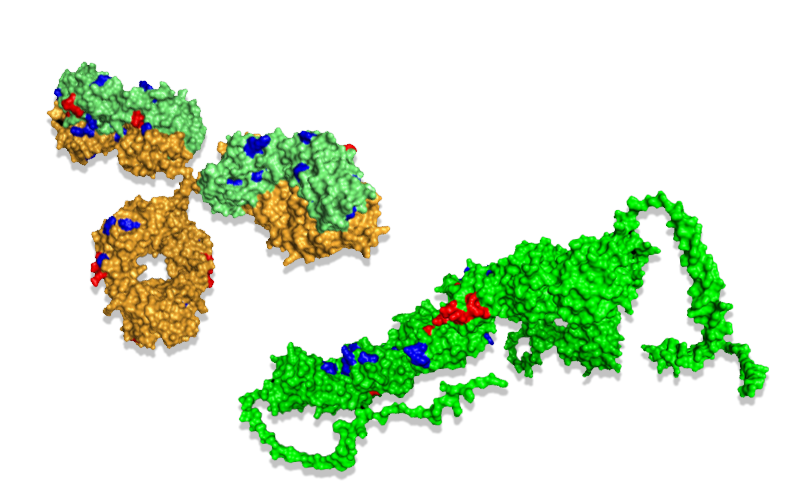
Mapping the oxidative labeling onto the antigen mCD163 (above) shows the epitope as decreased oxidation (red) and conformational changes as increased oxidation (blue).
Mapping the oxidative labeling onto the antibody shows both increased (blue) and decreased oxidation (red). The change in oxidation indicated both the paratope and dynamics that are in agreement with a previously reported closed form of IgG.[2]
Poster is presented at the American Society For Mass Spectrometry (ASMS) 2022 by Daniel Nyberg Larsen, PhD student from Ovodan Biotech and SDU.
[1] Graversen JH, Madsen M, Moestrup SK. CD163: a signal receptor scavenging haptoglobin-hemoglobin complexes from plasma. Int J Biochem Cell Biol. 2002 Apr;34(4):309-14. doi: 10.1016/s1357-2725(01)00144-3. PMID: 11854028.
[2] Maibom-Thomsen SL, Trier NH, Holm BE, Hansen KB, Rasmussen MI, Chailyan A, Marcatili P, Højrup P, Houen G. Immunoglobulin G structure and rheumatoid factor epitopes. PLoS One. 2019 Jun 14;14(6):e0217624. doi:
10.1371/journal.pone.0217624. PMID: 31199818; PMCID: PMC6568389.
3D structures created with BioRender.com